Dr. rer. nat. Urs Kindler

https://www.linkedin.com/in/dr-urs-kindler-074437228/
Technical Skills
- Programming Languages
- Python,
- R,
- Database Management
- SQL,
- Cypher (neo4J)
- Data Visualization:
- Tableau
Professional Experience
- Postdoctoral researcher at Leibniz Institute for Analytical Sciences - ISAS - e.V., Dortmund (2025 - now)
- Research Group Multidimensional Omics Data Analysis
- Scientific assistant at Ruhr University Bochum, (2019 - 2024)
- Department of Anatomy and Molecular Embryology
- Department of Human Genetics
- Scientific assistant at Max Delbrück Center for Molecular Medicine, Helmholtz Institute Berlin (2017 - 2019)
- Research Group of Cellular Neurosciences
Education
-
Data Analytics Bootcamp, ROGM Masterschool GmbH (2025)
-
Dr. rer. nat., Graduate School of Chemistry and Biochemistry, Ruhr University Bochum (2023)
-
M.Sc., International Master for Molecular and Developmental Stem Cell Biology, Ruhr University Bochum (2016)
-
B.Sc., Biology, Ruhr University Bochum (2014)
Projects
Academic Research Projects
Generation of Skeletal Muscle Organoids from Human Pluripotent Stem Cells
This project focused on developing and optimizing a protocol for generating 3D skeletal muscle organoids (SMOs) from human pluripotent stem cells (hESCs/hiPSCs). The goal was to create a model system that mimics human muscle development in vitro, allowing for the study of myogenesis and related diseases like muscular dystrophy.
Key Contributions:
- Protocol Development: Optimized the differentiation of hESCs/hiPSCs into SMOs hiPSCs.
- Cell Culture Expertise: Proficiently cultured human hiPSCs and applied the complex 3D differentiation protocol.
- Data Analysis & Visualization: Analyzed cell culture and differentiation data and created visualizations to present the results.
Skills Utilized:
- Cell culture
- 3D cell differentiation
- Data analysis
- Data visualization
- Protocol design and optimization
Why this project is important:
This project addressed the limited availability of muscle biopsies from patients with muscular dystrophy by providing a stable in vitro model for studying human myogenesis in healthy and diseased conditions.
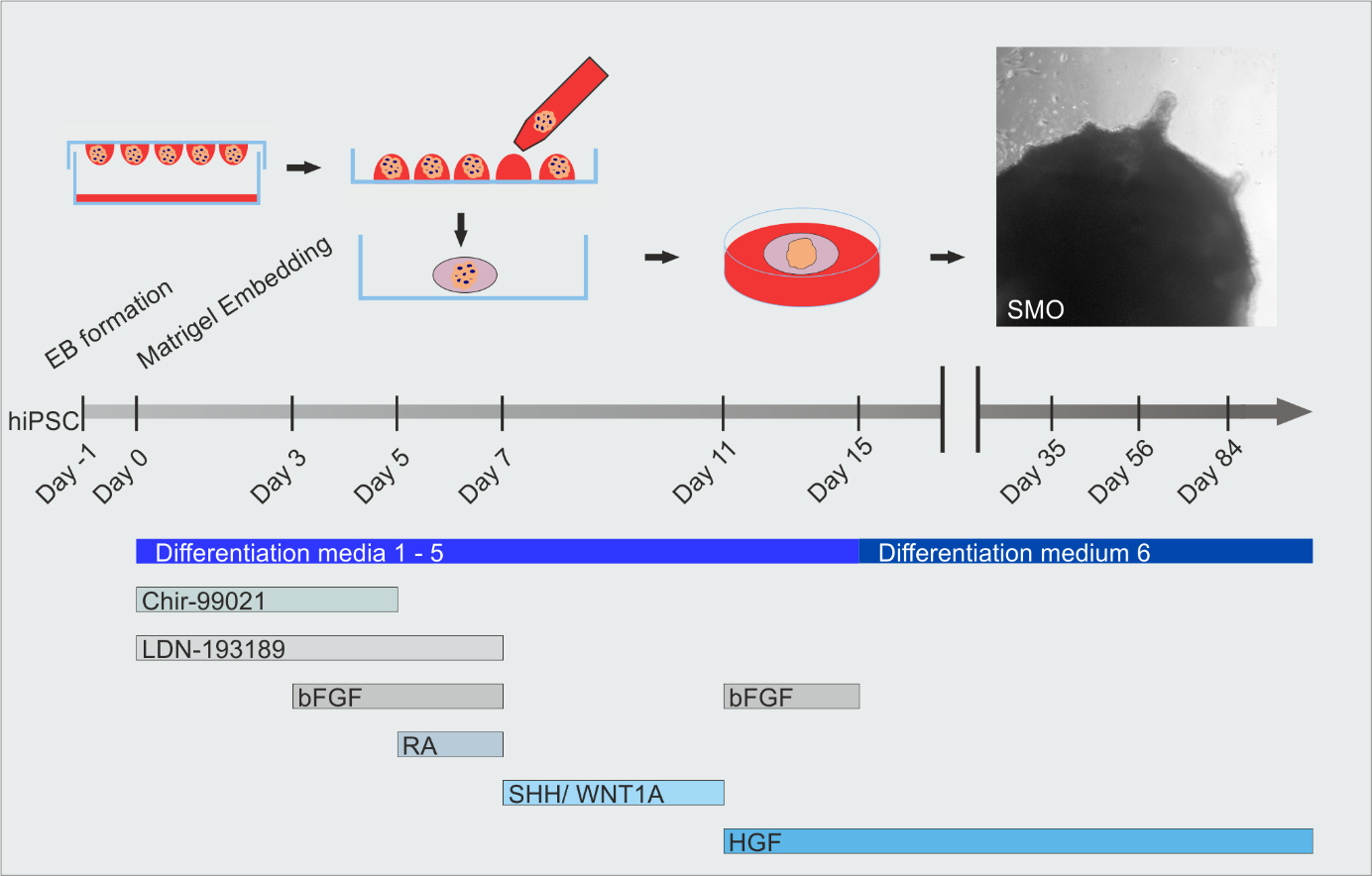
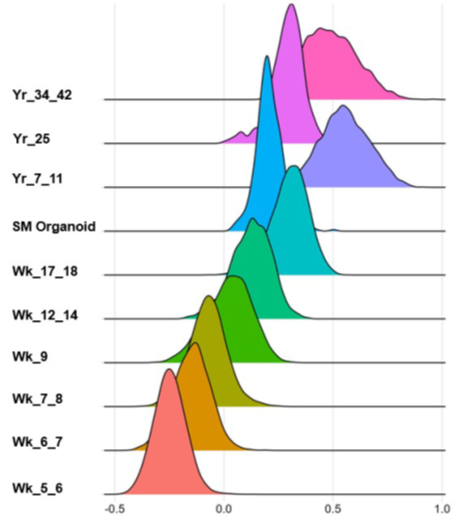
Human Skeletal Muscle Organoids Model Fetal Myogenesis and Sustain Uncommitted PAX7 Myogenic Progenitors
Myogenesis starts from early embryonic development until postnatal maturation. Until recently, little was known about the myogenic development in humans. Single cell analyses become more and more popular for the study of the heterogeneity of cell populations within one type of tissue. Upcoming scRNA-seq studies along different time points of muscle development start to clarify these ranges (Xi et al., 2020). Reconstruction of a developmental roadmap to investigate the maturation grade of myogenic progenitors along myogenic development gave an exact picture of in vitro differentiated cells as described by (Xi et al., 2020). Technically, to align the raw data to the human genome the Cell Ranger version 5 software was applied. To analyze organoid scRNA seq datasets, “Seurat” version 4.3 is used (Hao et al., 2021; Satija et al., 2015). The developmental score was calculated as described in (Xi et al., 2020). The ‘‘AddModuleScore’’ function was used to calculate the embryonic and adult score, using a list of differentially expressed genes (DEGs) between adult and embryonic myogenic progenitor clusters. The analysis were carried out using RStudio. A detailed description of the analysis and the 3D skeletal muscle organoid model can be found here (Kindler et al., 2024; Mavrommatis et al., 2023)
Data Science Projects:
Vehicle Category Prediction Using Machine Learning
Goal: Develop a machine learning model to predict unknown vehicle market categories and conduct comprehensive data analysis.
Description: This project involved analyzing a complex vehicle dataset and implementing a K-Nearest Neighbors (KNN) classification model to predict market categories. The focus was on data preprocessing, exploratory data analysis (EDA), and optimizing the model for better performance.
Data Visualization for Sales Analysis of an Emerging E-Commerce Business
This project involved creating an interactive Tableau dashboard to analyze sales data for Unicorn, a fictitious e-commerce company. The dataset simulated real-world business challenges faced by online retailers, offering insights into sales trends, regional performance, and product category dynamics. The dashboard served as a powerful tool for stakeholders to identify growth opportunities, address weak areas, and make data-driven decisions.
Goal: Develop a user-friendly Tableau dashboard to visualize key sales metrics, including revenue, profit, and product performance, and provide actionable recommendations for business growth.
Description: The project focused on building a dynamic and interactive Tableau dashboard that allowed stakeholders to explore sales data from 2015-2018. Tableau dashboard
Publication
Kindler, U., Mavrommatis, L., Käppler, F., Hiluf, D. G., Heilmann-Heimbach, S., Marcus, K., Günther Pomorski, T., Vorgerd, M., Brand-Saberi, B., & Zaehres, H. (2025). Duchenne Muscular Dystrophy Patient iPSCs—Derived Skeletal Muscle Organoids Exhibit a Developmental Delay in Myogenic Progenitor Maturation. Cells, 14(13), 1033. https://doi.org/10.3390/CELLS14131033/S1
Kindler, U., Zaehres, H., & Mavrommatis, L. (2024). Generation of Skeletal Muscle Organoids from Human Pluripotent Stem Cells. BIO-PROTOCOL, 14(1344). https://doi.org/10.21769/BIOPROTOC.4984
Mavrommatis, L., Zaben, A., Kindler, U., Kienitz, M.-C., Dietz, J., Jeong, H.-W., Böhme, P., Brand-Saberi, B., Vorgerd, M., & Zaehres, H. (2023). CRISPR/Cas9 genome editing in LGMD2A/R1 patient-derived induced pluripotent stem and skeletal muscle progenitor cells. Stem Cells International. https://doi.org/10.1155/2023/9246825
Mavrommatis L., Jeong H-W, Kindler U., Gomez-Giro G., Kienitz M-C., Stehling M., Psathaki OE., Zeuschner D., Bixel MG., Han D., Morosan-Puopolo G., Gerovska D., Yang JH., Kim JB, Araúzo-Bravo MJ, Schwamborn JC, Hahn SA, Adams RH, Schöler HR, Vorgerd M., Brand-Saberi B., Zaehres H. (2023). Human skeletal muscle organoids model fetal myogenesis and sustain uncommitted PAX7 myogenic progenitors. ELife, 12. https://doi.org/10.7554/ELIFE.87081
Brendel, M., Scharf, M., Kindler, U., Srirama, S., Divvela, K., & Brand-Saberi, B. (2023). Detection of Math6-Expressing Cell Types in Murine Placenta. Biology 2023, Vol. 12, Page 1252, 12(9), 1252. https://doi.org/10.3390/BIOLOGY12091252
Pan, Y., Xiong, M., Chen, R., Ma, Y., Corman, C., Maricos, M., Kindler, U., Semtner, M., Chen, Y.-H., Dahiya, S., & Gutmann, D. H. (2018). Athymic mice reveal a requirement for T-cell-microglia interactions in establishing a microenvironment supportive of Nf1 low-grade glioma growth. Genes and Development, 32(7–8). https://doi.org/10.1101/gad.310797.117